|
chrX_-_63450486
|
0.682
|
NM_130388
|
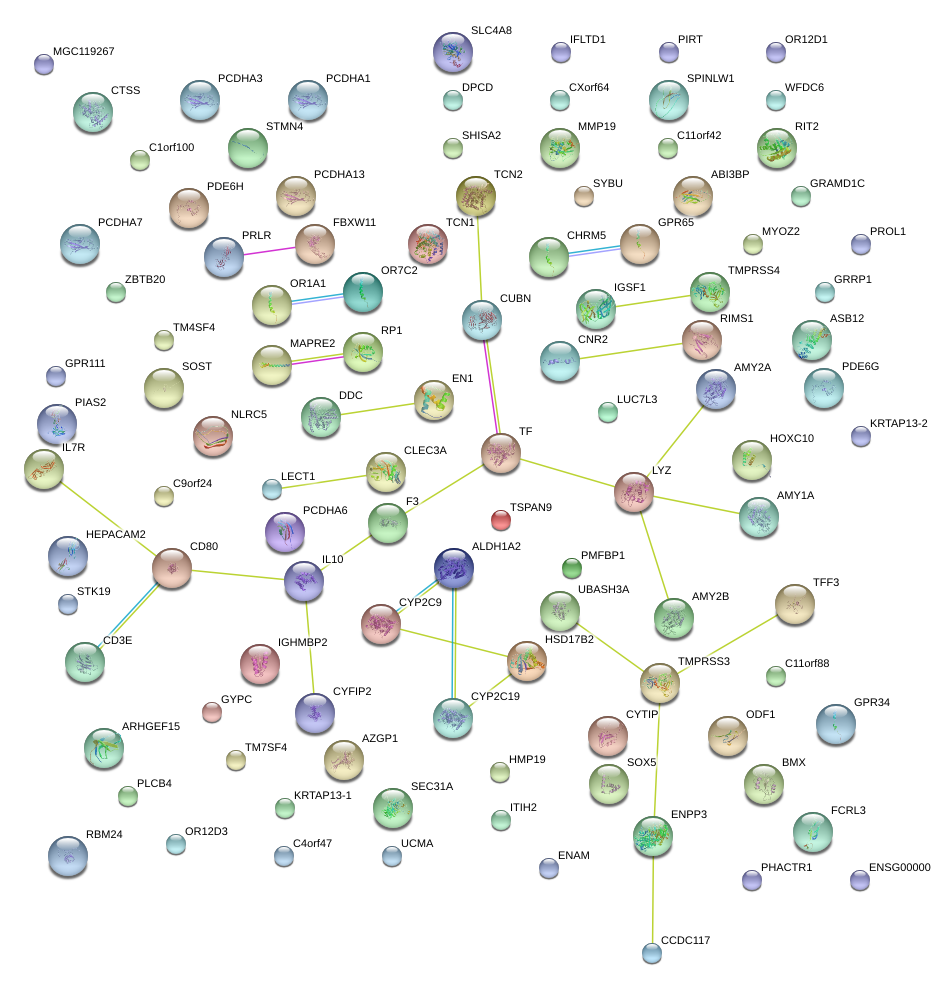
ASB12
|
ankyrin repeat and SOCS box containing 12
|
|
chr8_-_27115935
|
0.593
|
|
STMN4
|
stathmin-like 4
|
|
chr21_-_43735462
|
0.580
|
|
TFF3
|
trefoil factor 3 (intestinal)
|
|
chr3_+_133495925
|
0.548
|
|
TF
|
transferrin
|
|
chr10_+_7745341
|
0.544
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr10_+_7745354
|
0.542
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr10_-_13276327
|
0.519
|
NM_145314
|
UCMA
|
upper zone of growth plate and cartilage matrix associated
|
|
chr10_+_7745325
|
0.507
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr5_+_140165875
|
0.474
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr7_-_99573637
|
0.471
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chr21_+_43823970
|
0.447
|
NM_001001895
NM_001243467
NM_018961
|
UBASH3A
|
ubiquitin associated and SH3 domain containing A
|
|
chr2_-_119605758
|
0.440
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr12_-_25706177
|
0.438
|
NM_152590
NM_001145728
NM_001145729
|
IFLTD1
|
intermediate filament tail domain containing 1
|
|
chr12_+_69742130
|
0.431
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr10_+_96522457
|
0.429
|
NM_000769
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19
|
|
chr20_-_44175995
|
0.422
|
NM_001198986
NM_020398
|
SPINLW1-WFDC6
SPINLW1
|
SPINLW1-WFDC6 readthrough
serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin)
|
|
chr10_+_7745229
|
0.413
|
NM_002216
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr4_+_71494460
|
0.407
|
NM_031889
|
ENAM
|
enamelin
|
|
chr20_+_9198036
|
0.403
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr12_+_69742140
|
0.396
|
|
LYZ
|
lysozyme
|
|
chr5_+_156820161
|
0.394
|
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr21_+_31768294
|
0.385
|
NM_181599
|
KRTAP13-1
|
keratin associated protein 13-1
|
|
chrX_+_15525430
|
0.372
|
NM_001721
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr4_+_186350543
|
0.365
|
NM_001114357
|
C4orf47
|
chromosome 4 open reading frame 47
|
|
chr9_-_34397786
|
0.363
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr4_+_120056938
|
0.361
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr12_+_15125955
|
0.352
|
NM_006205
|
PDE6H
|
phosphodiesterase 6H, cGMP-specific, cone, gamma
|
|
chr5_+_173472692
|
0.350
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chrX_+_41548225
|
0.349
|
NM_001097579
NM_005300
|
GPR34
|
G protein-coupled receptor 34
|
|
chr10_+_7745284
|
0.338
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr5_+_140213968
|
0.331
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr5_-_35230548
|
0.323
|
|
PRLR
|
prolactin receptor
|
|
chr1_+_244515936
|
0.317
|
NM_001012970
|
C1orf100
|
chromosome 1 open reading frame 100
|
|
chr8_-_110620220
|
0.315
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr22_+_31003069
|
0.308
|
NM_000355
NM_001184726
|
TCN2
|
transcobalamin II
|
|
chr11_+_118175294
|
0.296
|
NM_000733
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex)
|
|
chr15_-_58306185
|
0.295
|
NM_170697
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr13_-_53313924
|
0.285
|
NM_001011705
NM_007015
|
LECT1
|
leukocyte cell derived chemotaxin 1
|
|
chr6_+_47624222
|
0.257
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr1_-_150738255
|
0.254
|
|
CTSS
|
cathepsin S
|
|
chr13_-_26625164
|
0.251
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr16_+_78062985
|
0.239
|
NM_001244755
|
CLEC3A
|
C-type lectin domain family 3, member A
|
|
chr21_-_43735705
|
0.236
|
NM_003226
|
TFF3
|
trefoil factor 3 (intestinal)
|
|
chr2_+_127413739
|
0.236
|
|
GYPC
|
glycophorin C (Gerbich blood group)
|
|
chr8_-_27115897
|
0.225
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr16_+_82068958
|
0.224
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr2_-_158300517
|
0.223
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr14_+_88471467
|
0.209
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr11_+_111385509
|
0.207
|
NM_001100388
NM_207430
|
C11orf88
|
chromosome 11 open reading frame 88
|
|
chr6_+_12717036
|
0.202
|
NM_001242648
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr7_-_92848837
|
0.198
|
NM_198151
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr4_+_71263598
|
0.197
|
NM_021225
|
PROL1
|
proline rich, lacrimal 1
|
|
chr11_+_6226797
|
0.196
|
NM_173525
|
C11orf42
|
chromosome 11 open reading frame 42
|
|
chr1_+_104159953
|
0.188
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chr17_+_48821106
|
0.187
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr10_-_17171789
|
0.186
|
NM_001081
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr5_+_35856990
|
0.184
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr17_-_41836132
|
0.184
|
NM_025237
|
SOST
|
sclerostin
|
|
chrX_-_130423199
|
0.178
|
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chr12_-_56236614
|
0.177
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr3_-_114343791
|
0.176
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr19_+_15052298
|
0.176
|
NM_012377
|
OR7C2
|
olfactory receptor, family 7, subfamily C, member 2
|
|
chr11_-_59633942
|
0.171
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chr16_+_57050985
|
0.166
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr6_+_131958441
|
0.160
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr16_-_72206023
|
0.158
|
NM_031293
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr20_-_44168128
|
0.156
|
NM_080827
|
WFDC6
|
WAP four-disulfide core domain 6
|
|
chr10_+_96698409
|
0.156
|
NM_000771
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9
|
|
chr8_+_103563847
|
0.154
|
NM_024410
|
ODF1
|
outer dense fiber of sperm tails 1
|
|
chr21_-_43816123
|
0.152
|
NM_024022
NM_032405
|
TMPRSS3
|
transmembrane protease, serine 3
|
|
chr7_-_50633042
|
0.152
|
NM_001082971
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr20_+_9494995
|
0.147
|
NM_001199897
NM_012261
|
LAMP5
|
lysosomal-associated membrane protein family, member 5
|
|
chr17_-_10741398
|
0.147
|
NM_001101387
|
PIRT
|
phosphoinositide-interacting regulator of transient receptor potential channels
|
|
chr17_+_3118911
|
0.146
|
NM_014565
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1
|
|
chr12_-_24103953
|
0.142
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr1_-_206945725
|
0.141
|
NM_000572
|
IL10
|
interleukin 10
|
|
chr3_-_100712248
|
0.140
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr1_+_26485510
|
0.139
|
NM_024869
|
FAM110D
|
family with sequence similarity 110, member D
|
|
chr7_-_50628744
|
0.137
|
NM_000790
NM_001242886
NM_001242887
NM_001242888
NM_001242889
NM_001242890
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr1_-_157670280
|
0.136
|
NM_052939
|
FCRL3
|
Fc receptor-like 3
|
|
chr5_+_140180782
|
0.135
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr17_+_8214139
|
0.135
|
NM_025014
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr15_+_34261088
|
0.134
|
NM_012125
|
CHRM5
|
cholinergic receptor, muscarinic 5
|
|
chr6_+_17282931
|
0.129
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr18_-_40695483
|
0.128
|
NM_002930
|
RIT2
|
Ras-like without CAAX 2
|
|
chr1_-_24239758
|
0.127
|
NM_001841
|
CNR2
|
cannabinoid receptor 2 (macrophage)
|
|
chr3_+_149192479
|
0.127
|
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr3_-_119278402
|
0.126
|
NM_005191
|
CD80
|
CD80 molecule
|
|
chr3_+_113616317
|
0.124
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chrX_+_125953746
|
0.123
|
NM_001122716
|
CXorf64
|
chromosome X open reading frame 64
|
|
chr6_-_29343063
|
0.122
|
NM_030959
|
OR12D3
|
olfactory receptor, family 12, subfamily D, member 3
|
|
chr5_-_35230822
|
0.121
|
NM_000949
|
PRLR
|
prolactin receptor
|
|
chr8_+_105352050
|
0.120
|
NM_030788
|
TM7SF4
|
transmembrane 7 superfamily member 4
|
|
chr12_+_54378929
|
0.120
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr6_+_72926144
|
0.119
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr8_+_55528626
|
0.118
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chr1_+_84768333
|
0.117
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr1_-_217250230
|
0.117
|
NM_001243507
NM_001243509
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr2_+_150187028
|
0.114
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr19_+_15852200
|
0.113
|
NM_013938
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3
|
|
chr19_-_14992166
|
0.113
|
NM_030901
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17
|
|
chr3_-_93692592
|
0.112
|
|
PROS1
|
protein S (alpha)
|
|
chr3_+_108541544
|
0.111
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr2_+_69201704
|
0.111
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr1_+_120049825
|
0.110
|
NM_000862
|
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1
|
|
chr4_-_84035888
|
0.109
|
|
PLAC8
|
placenta-specific 8
|
|
chr11_-_61124223
|
0.108
|
NM_001161452
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr11_+_111789658
|
0.107
|
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr3_+_149192367
|
0.107
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr6_-_152702329
|
0.107
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr9_-_135819953
|
0.107
|
|
TSC1
|
tuberous sclerosis 1
|
|
chr16_-_53737713
|
0.106
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr1_-_151148425
|
0.105
|
NM_013353
|
TMOD4
|
tropomodulin 4 (muscle)
|
|
chr15_-_90349819
|
0.104
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chrX_+_37639306
|
0.101
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr14_-_94854910
|
0.101
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr1_-_202129731
|
0.101
|
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr11_+_15136483
|
0.101
|
NM_001042536
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chr14_-_94855120
|
0.100
|
NM_000295
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr2_+_102927961
|
0.100
|
NM_016232
|
IL18R1
IL1RL1
|
interleukin 18 receptor 1
interleukin 1 receptor-like 1
|
|
chr20_-_33460558
|
0.100
|
NM_178026
|
GGT7
|
gamma-glutamyltransferase 7
|
|
chr22_+_51176623
|
0.099
|
NM_001097
|
ACR
|
acrosin
|
|
chr20_+_2276612
|
0.098
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr2_+_191002485
|
0.098
|
NM_001042521
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr5_+_38445577
|
0.098
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr1_-_26644755
|
0.098
|
NM_145345
|
UBXN11
|
UBX domain protein 11
|
|
chr2_-_49381555
|
0.097
|
NM_000145
NM_181446
|
FSHR
|
follicle stimulating hormone receptor
|
|
chr11_+_111789600
|
0.097
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr8_-_86290333
|
0.097
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr4_-_84035908
|
0.096
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr13_+_52436116
|
0.096
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr19_+_55476651
|
0.092
|
NM_001174081
|
NLRP2
|
NLR family, pyrin domain containing 2
|
|
chr7_+_28475233
|
0.091
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_+_148036528
|
0.091
|
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr3_-_15642969
|
0.090
|
|
HACL1
|
2-hydroxyacyl-CoA lyase 1
|
|
chr1_+_244214576
|
0.090
|
|
ZNF238
|
zinc finger protein 238
|
|
chr6_+_7108184
|
0.089
|
|
RREB1
|
ras responsive element binding protein 1
|
|
chr15_+_76352270
|
0.089
|
NM_152335
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chr6_-_52710892
|
0.089
|
NM_153699
|
GSTA5
|
glutathione S-transferase alpha 5
|
|
chr2_-_190927321
|
0.088
|
NM_005259
|
MSTN
|
myostatin
|
|
chr18_-_56296188
|
0.087
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr5_+_72469022
|
0.087
|
NM_153217
|
TMEM174
|
transmembrane protein 174
|
|
chr14_+_29236882
|
0.085
|
|
FOXG1
|
forkhead box G1
|
|
chr4_+_160188997
|
0.085
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr9_-_104198045
|
0.085
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr12_+_52306112
|
0.083
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr11_-_9225780
|
0.082
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr16_+_82068830
|
0.082
|
NM_002153
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr12_+_100867550
|
0.081
|
NM_001206977
NM_001206978
NM_001206979
NM_005123
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr1_-_217113007
|
0.081
|
NM_001243512
NM_001243513
NM_001243510
NM_001243511
|
ESRRG
|
estrogen-related receptor gamma
|
|
chrM_+_9725
|
0.081
|
|
|
|
|
chr11_+_59824054
|
0.081
|
NM_001031666
NM_001031809
NM_006138
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific)
|
|
chr5_-_156593178
|
0.080
|
NM_130899
|
FAM71B
|
family with sequence similarity 71, member B
|
|
chr10_-_112678902
|
0.079
|
NM_001195304
NM_001195305
NM_001195307
NM_001243783
|
BBIP1
|
BBSome interacting protein 1
|
|
chr4_+_25657434
|
0.079
|
NM_001177998
NM_006424
|
SLC34A2
|
solute carrier family 34 (sodium phosphate), member 2
|
|
chr3_+_185046682
|
0.078
|
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr5_+_140474198
|
0.078
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr9_+_124048396
|
0.078
|
|
GSN
|
gelsolin
|
|
chr6_+_30908776
|
0.077
|
NM_080870
|
DPCR1
|
diffuse panbronchiolitis critical region 1
|
|
chrX_+_68835910
|
0.077
|
NM_001005609
NM_001005610
NM_001005612
NM_001005613
NM_001399
|
EDA
|
ectodysplasin A
|
|
chr4_-_122854192
|
0.077
|
NM_003305
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr6_-_29055048
|
0.077
|
NM_001005226
|
OR2B3
|
olfactory receptor, family 2, subfamily B, member 3
|
|
chr7_-_55930444
|
0.076
|
NM_207366
|
SEPT14
|
septin 14
|
|
chr3_-_15643063
|
0.075
|
NM_012260
|
HACL1
|
2-hydroxyacyl-CoA lyase 1
|
|
chr6_-_155777036
|
0.074
|
NM_015718
|
NOX3
|
NADPH oxidase 3
|
|
chr5_-_39424930
|
0.074
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr5_+_173472737
|
0.074
|
|
HMP19
|
HMP19 protein
|
|
chr14_+_76071804
|
0.074
|
NM_001195283
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr1_-_36948423
|
0.073
|
|
CSF3R
|
colony stimulating factor 3 receptor (granulocyte)
|
|
chr17_-_3794036
|
0.073
|
NM_172206
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha
|
|
chr17_-_3599298
|
0.072
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr17_-_36105005
|
0.070
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr4_-_100140330
|
0.070
|
NM_000672
NM_001102470
|
ADH6
|
alcohol dehydrogenase 6 (class V)
|
|
chrX_+_74493893
|
0.070
|
NM_145052
|
UPRT
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae)
|
|
chr11_+_62009723
|
0.069
|
NM_006551
|
SCGB1D2
|
secretoglobin, family 1D, member 2
|
|
chr17_-_53800074
|
0.069
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chrX_+_37639246
|
0.069
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr5_+_176873795
|
0.068
|
NM_001174101
NM_030567
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr6_+_123100860
|
0.068
|
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr7_-_87856282
|
0.067
|
NM_198901
|
SRI
|
sorcin
|
|
chr10_-_104178569
|
0.067
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr1_+_163038934
|
0.066
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr14_-_24777064
|
0.066
|
|
|
|
|
chr3_-_116164363
|
0.065
|
NM_002338
|
LSAMP
|
limbic system-associated membrane protein
|
|
chr6_+_28109611
|
0.064
|
NM_006298
|
ZNF192
|
zinc finger protein 192
|
|
chr6_+_22569656
|
0.064
|
NM_138574
|
HDGFL1
|
hepatoma derived growth factor-like 1
|
|
chr2_+_179184970
|
0.064
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr4_+_36283243
|
0.064
|
NM_001136536
|
DTHD1
|
death domain containing 1
|
|
chr17_-_79623561
|
0.064
|
NM_002602
|
PDE6G
|
phosphodiesterase 6G, cGMP-specific, rod, gamma
|
|
chr2_+_33701678
|
0.063
|
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr11_-_5255712
|
0.062
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr3_-_42917191
|
0.062
|
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr1_+_27719151
|
0.062
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr16_+_69958900
|
0.062
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr15_-_42186274
|
0.061
|
NM_016642
|
SPTBN5
|
spectrin, beta, non-erythrocytic 5
|
|
chr20_+_20037289
|
0.061
|
NM_001167816
|
C20orf26
|
chromosome 20 open reading frame 26
|
|
chr5_+_140201221
|
0.060
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr17_-_47439326
|
0.060
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr11_+_65271145
|
0.060
|
|
|
|
|
chr1_-_203155910
|
0.060
|
NM_001276
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|